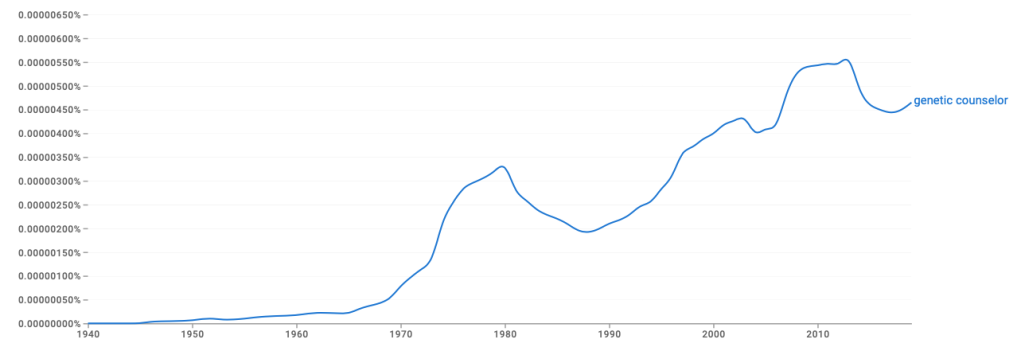
A guest post by Ambreen Khan and Kimberly Zayhowski
Normative standards of professionalism dictate that a professional should remain apolitical, positing that separating personal beliefs from professional endeavors allows one to maintain objectivity. The enforcement of these standards is increasingly evident in genetic counseling spaces, such as with censorship in workplace meetings, on discussion forums and social media, and at conferences.
However, remaining apolitical grows complex given the politicization of everyone’s identities and personhood. The intertwining of personal, political, and professional realms is undeniable, often operating subconsciously. Operating in a makeshift bubble of neutrality disconnects us from the lived realities of our colleagues and the patients we strive to serve.
Eugenics underpinnings in genetic counseling
The roots of the genetic counseling profession are entangled with a history steeped in eugenics, a movement advocating for selective breeding to enhance the human population by using erroneous assumptions about genetics shaped primarily by social, political and personal biases of its supporters. Originating in the late 19th century, eugenics principles guided the atrocities committed during Nazi Germany’s reign, heavily shaped by American eugenicists like Charles Davenport and studies from the Eugenics Record Office at Cold Spring Harbor Laboratory. The historical justification of eugenics to forcibly sterilize, criminalize, and perpetrate genocide against minoritized communities has been rooted in the misappropriation of genetic and medical concepts. Therefore, dismissing the importance of politics in the field of genetics is a fallacy.
Genetic counseling’s origins can be traced back to the ethically fraught ideology of breeding out those considered less “desirable.” The justification for establishing and funding the first genetic counseling program suggested that genetic counseling serves as a strategy to mitigate hereditary diseases and encourage individuals to make informed decisions regarding reproduction, both for their own well-being and that of the broader population.
Despite the prevailing belief among genetic counselors that we are staunchly anti-eugenics, traces of eugenic ideology persist within certain aspects of our practice, aligning with broader political and power structures. This is exemplified by the recent NSGC Practice Guidelines suggesting the use of expanded carrier screenings as a means for downstream cost-savings through the prevention of births of individuals with certain genetic conditions.
Moreover, genetic counselors’ desire for absolute neutrality ties closely with the need to adopt a non-directive approach with patients, obscuring the intrinsically directive nature of everything said and done in patient interactions. This connection can be traced back to post-WWII geneticists’ efforts to distance themselves from eugenics, despite perpetuating comparable ideologies under the guise of neutrality. The norm to remain apolitical perpetuates self-censorship, impeding the field’s ability to openly confront its connections with eugenics.
The impossibility of neutrality
Acknowledging personal political beliefs becomes a crucial aspect of a genetic counselor’s professional journey and our interactions with colleagues. As Lewis Wallace, a transgender reporter, asserts in his piece titled “Objectivity Is Dead And I’m Okay With It,” neutrality is not real, particularly for people with marginalized identities who cannot remain neutral or centrist in debates concerning their own humanity. Hence, the structures demanding neutrality in the face of oppression must be challenged.
Numerous contemporary policies, such as those regarding immigration, disability and reproductive rights, racial justice, education, LGBTQIA+ rights, colonialism, imperialism, and more, directly impact how we show up in our professional lives. Policies can impede our capacity to pursue a career, such as when they impact visa status, restrict access to safe and inclusive work environments, or perpetuate discriminatory practices such as anti-transgender legislation.
The politicization of religious and ethnic identities to justify violence exposes individuals to bigotry, threatening their safety and sense of belonging. Politics can profoundly affect mental and physical well-being, as evidenced by US-funded genocide in Gaza and settler colonial violence in the occupied West Bank, leading to distress and safety concerns among Palestinian genetic counselors as well as allies that speak out against these atrocities. In such instances, neutrality serves oppressors, demanding marginalized individuals to suppress their emotions and well-being to conform to “professionalism” standards, which expect silence amid oppression.
The burden of representation and palatability
Standards of professionalism carry oppressive ideologies favoring white supremacy. Professionalism traditionally reflects the cultural norms, behaviors, and traits of the dominant social group, often represented by straight, cisgender, non-disabled white men in the broader field of medicine in the US, or women in genetic counseling. Consequently, professionalism tends to be assessed primarily among those who are racially minoritized, queer, gender-diverse, and disabled. An expectation of professionalism entails the ability to collaborate with others – even if those people say and do awful things. This creates an environment of dealing with microaggressions and discrimination quietly and laying low when witnessing bigoted conversations.
In navigating political dynamics, genetic counselors often encounter challenges with colleagues tied to respectability politics, a phenomenon where individuals from marginalized groups feel compelled to conform to mainstream expectations to gain social acceptance. Additionally, the pitfalls of “whataboutisms” arise, deflecting from the core issues at hand by pointing to separate problems or situations. This tactic often undermines meaningful discussions about systemic problems, diverting attention from the pressing need for change.
The pursuit of “palatability” within diversity, equity, and inclusion work can paradoxically prioritize the comfort of the oppressor over meaningful progress. Efforts to make conversations or initiatives more palatable risk diluting the urgency and discomfort inherent in addressing systemic issues. Individuals with minoritized identities often find themselves assuming the role of ambassadors for their communities. As they navigate professional spaces, they become de facto representatives, sharing the responsibility of dispelling stereotypes and fostering understanding. This burden is a consequence of existing in spaces where diversity is limited.
Integrating our personal, political, and professional selves
True progress in social justice work demands confronting challenging truths, dismantling entrenched power structures, and prioritizing the voices of marginalized communities over the comfort of those with privilege. Achieving this necessitates a deep understanding of one’s own privileges through an intersectional lens.
Without reflecting on the underlying reasons that necessitate our need to maintain objectivity while upholding the status quo, genetic counselors jeopardize their ability to engage in nuanced conversations with colleagues and patients. Staying engaged in global affairs is essential for genetic counselors to confront their personal biases and improve patient care.
Trusting ourselves and our colleagues to bring their authentic, political selves to professional spaces promotes meaningful dialogue and mutual understanding. The myth of apolitical neutrality acts as a barrier to recognizing the complexity of human experiences among colleagues and within ourselves. As we navigate the paradox of remaining apolitical in a world where identities are inherently political, genetic counselors must consistently question the root cause of their need for neutrality.
The opinions expressed in this article are solely our own and do not reflect the views and opinions of our employers.
Authors:
*Ambreen Khan, MS, CGC (she/her) works as a laboratory genetic counselor and a grassroots community organizer. A bilingual Muslim individual of Pakistani descent, Ambreen follows her passion of increasing access to equitable genetic services locally and globally, through educational talks, social media content, and research.
*Kimberly Zayhowski, MS, CGC (she/her) works as an assistant professor and research genetic counselor. A queer and multiracial individual, Kim is dedicated to advocating against oppression in genetic counseling research, education, and practice.
*Names in alphabetical order. These authors have contributed equally to this work.